Decode 100% of the Genome with AI
We transform noisy data from whole genome sequencing into actionable insights covering coding and non-coding DNA.
Cutting-edge technology and precision at scale for research, drug discovery, and clinical diagnostics.
A spin-off from:
Published in:
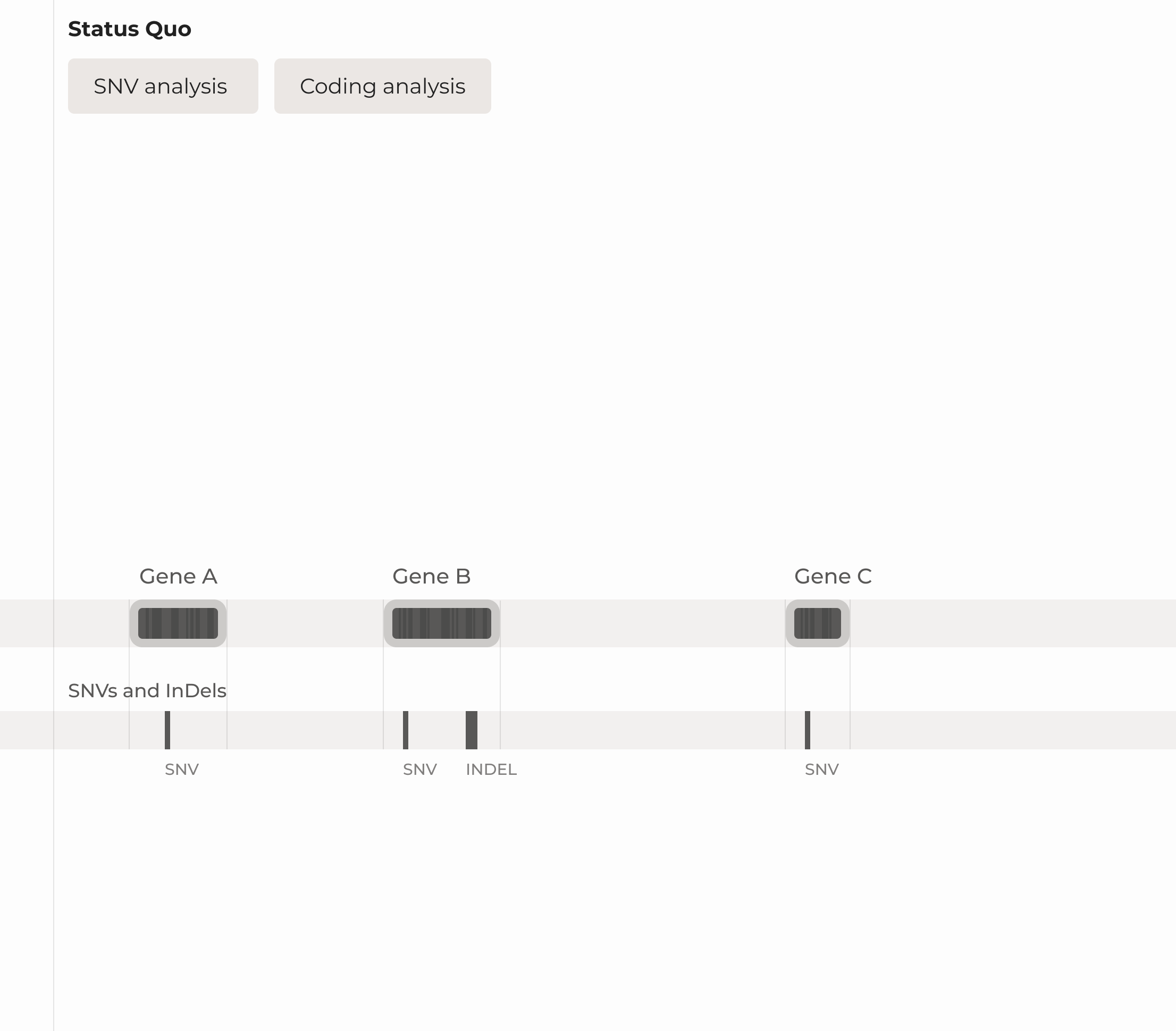
All-in-one AI powered platform for variant detection, prioritization, interpretation and data sharing
100% Genome Coverage
Fast Large-Scale Cohort Analysis
81% Fewer False Positives
Complete Variant Detection
Full Multi-Omics Integration
What we offer
Comprehensive genome analysis
Our software analyses the whole genome on cohort, family or individual level tailored to your needs.
Variation detection and ranking
Through the detection, ranking, and prioritization of mutations, you are provided with a manageable list of suggested pathogenic mutations.
Cutting edge AI Technology
Detection
High-quality detection methods that decrease false-positive structural variants by 81%, including DEL, DUP, INS and INV.
Prioritization
Disease-agnostic AI tool ranks over 80% of known pathogenic variants in the top 10. On top of that, we created a disease-specific scoring system with over 90% accuracy, it ensures precise prioritization for more reliable research outcomes.
Discover our Platform
Get a personalized walkthrough of our platform and see how it can benefit your needs.
Resources
Lucid Genomics at ASHG 2025 conference and industry session
2025 October 16
We were at ASHG 2025 with our booth and Professor Pawel Stankiewicz from Baylor College of Medicine lead our Industry Education Session showcasing how non-coding and structural variants reveal disease mechanisms, powered by Lucid’s AI platform
Lucid Genomics at BioTechX 2025 conference
2025 October 10
We participated in BioTechX Europe engaging with biotech and pharma leaders on how we can support the detection and interpretation of structural variants to accelerate biomarker discovery and patient stratification in drug discovery
The 100 most important minds in Berlin science 2025
2025 September 30
Our CEO Uirá Souto Melo and CTO M-Hossein Moeinzadeh have been recognized among Tagesspiegel’s “100 most important minds in Berlin science 2025.” The recognition highlights how Lucid Genomics is bringing the non-coding genome into mainstream awareness.
Stay informed
Stay informed with the latest updates, news, and insights delivered to your inbox.

















