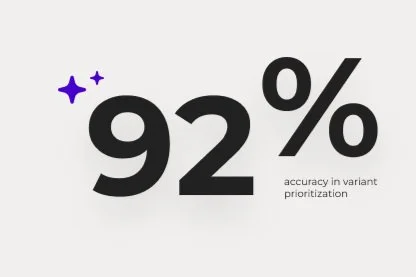
Harness deep learning and AI to achieve up to 92% accuracy in variant prioritization for faster, more reliable research.
Proof of Concept will be published in Q1 2025
Detection
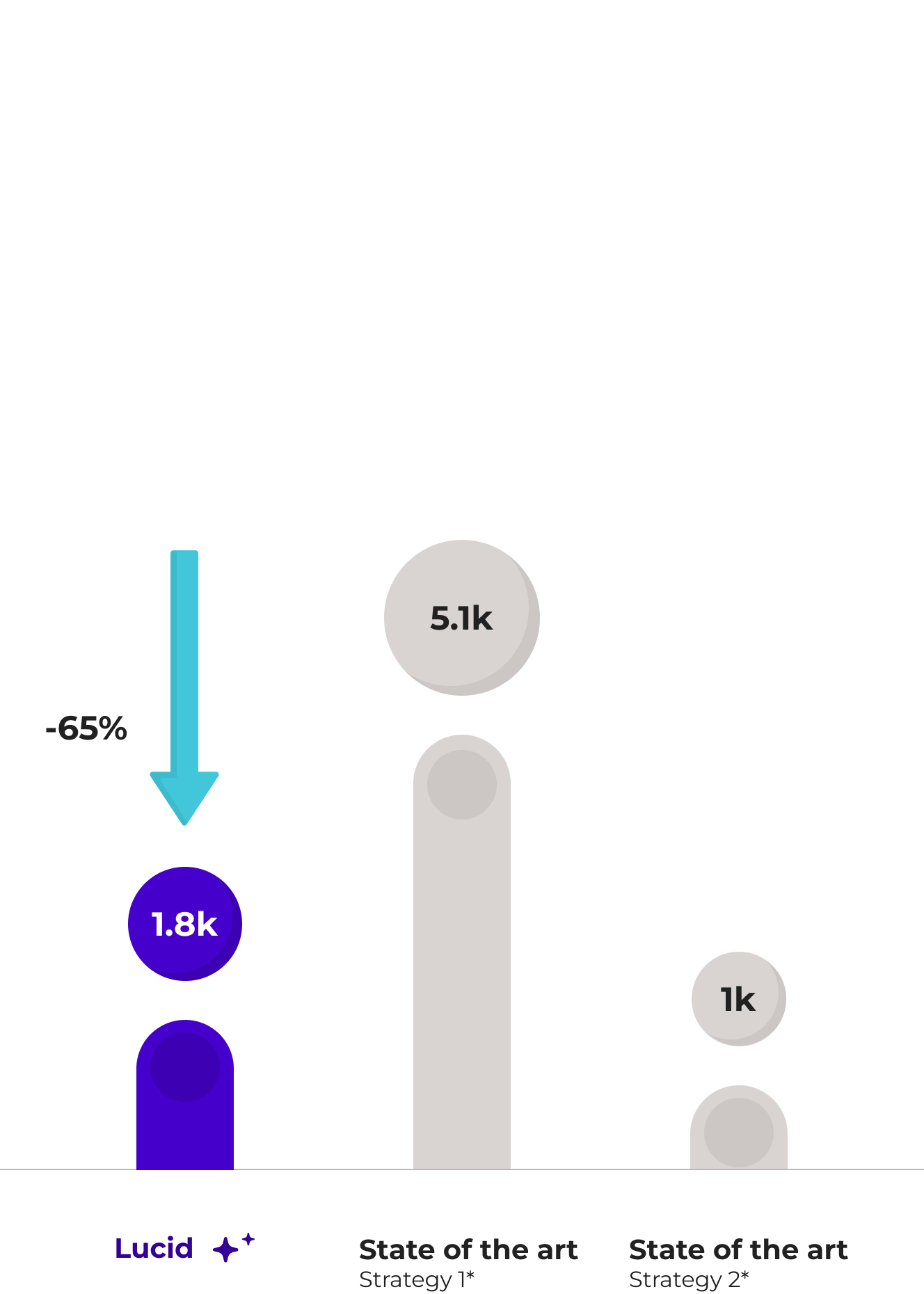
We developed an AI model to confidently identify SVs (DEL, DUP, INS and INV) across the entire genome, even with noisy short-read sequencing data. Our model reduces the number of false positives by up to 65% given the union of state-of-the-art SV callers while increasing the number of true positives by 200% compared to conventionally used consensus approaches.
True positives
Number of structural variants
False positives
Number of structural variants
*1: Union of 5 state-of-the-art SV callers
*2: Commonly employed consensus strate*1: Union of 5 state-of-the-art SV callersgies (at least 2/5 Callers)
Prioritization
With TADA, our disease-agnostic AI method, we place 18% more known pathogenic SVs among the top 10 candidates compared to best-in-class open-source algorithms.
Incorporating disease-specific regulatory information in the Lucid expert decision system boosts our performance even further to more than 90% for selected diseases
Start now!
Get a personalized walkthrough of our platform and see how it can benefit your needs.
Learn more about our research
AML with complex karyotype: extreme genomic complexity revealed by combined long-read sequencing and Hi-C technology
2024 November. 10
Acute myeloid leukemia with complex karyotype (CK-AML) is associated with poor prognosis, which is only in part explained by underlying TP53 mutations.
Enhancer hijacking at the ARHGAP36 locus is associated with connective tissue to bone transformation
2023 April. 11
Heterotopic ossification is a disorder caused by abnormal mineralization of soft tissues in which signaling pathways such as BMP, TGFβ, and WNT are known key players in driving ectopic bone formation.
TADA - A machine learning tool for functional annotation-based prioritisation of pathogenic CNVs
2022 March. 01
Few methods have been developed to investigate copy number variants (CNVs) based on their predicted pathogenicity. We introduce TADA, a method to prioritise pathogenic CNVs through …